Drug design, drug delivery and technologies, BioWorld Science
Drug Design, Drug Delivery & Technologies
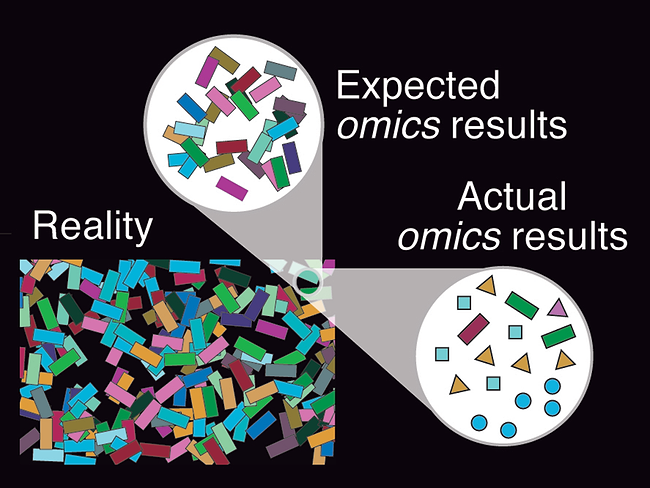
Incomplete databases can yield false positives in microbiome studies
Read MoreDrug Design, Drug Delivery & Technologies
Model transforms zinc finger design into 'push-button' technology
Read MoreDrug Design, Drug Delivery & Technologies
Genkore establishes collaboration on in vivo gene editing therapies
Read MoreDrug Design, Drug Delivery & Technologies